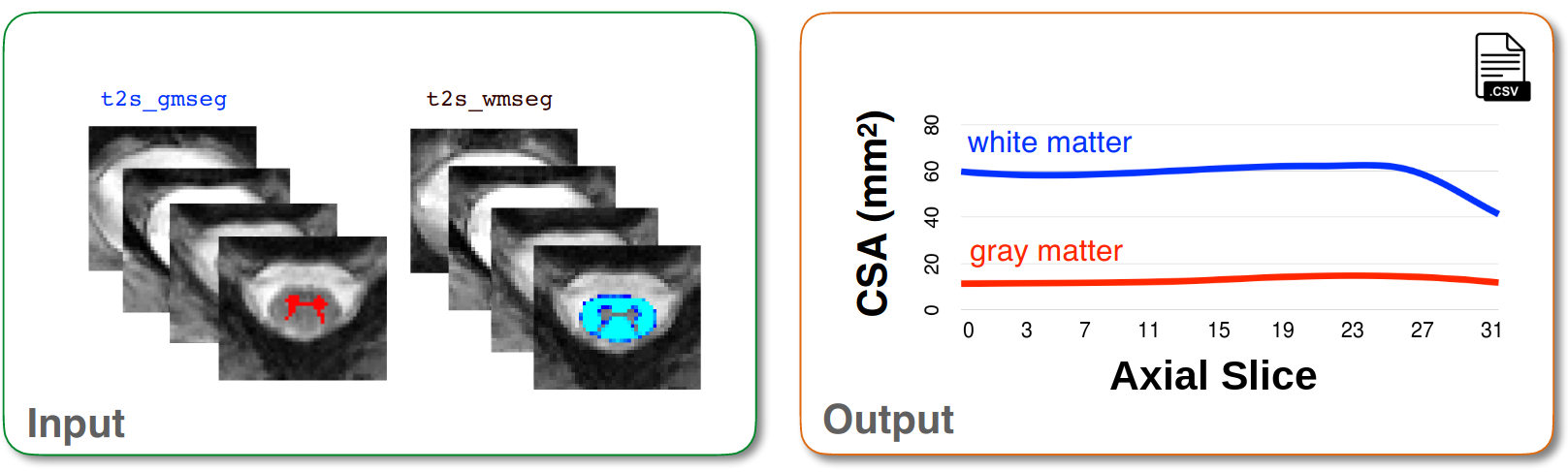
Using binary masks to compute CSA for gray and white matter¶
First, we will use the gray and white matter segmentations to compute the cross sectional area of GM and WM. This is achieved using sct_process_segmentation.
Important
There is a limit to the precision you can achieve for a given image resolution. SCT does not truncate spurious digits when performing angle correction, so please keep in mind that there may be non-significant digits in the computed values. You may wish to compare angle-corrected values with their corresponding uncorrected values to get a sense of the limits on precision.
Compute CSA¶
sct_process_segmentation -i t2s_wmseg.nii.gz -o csa_wm.csv -perslice 1 -angle-corr 0
sct_process_segmentation -i t2s_gmseg.nii.gz -o csa_gm.csv -perslice 1 -angle-corr 0
- Input arguments:
-i: The input segmentation file.-o: The output CSV file.-perslice: Set this option to 1 to turn on per-slice computation.-angle-corr 0: Normally, angle correction will be applied duringsct_process_segmentationto account for scans where the spinal cord is positioned at an angle with respect to the superior-inferior axis. While this works well when the input is a full spinal cord segmentation, in this case we are instead providing GM/WM segmentations only. The shape of these segmentations can have a negative effect on so the estimation of the cord centerline, which in turn may cause the estimated angle to be incorrect. So, here we specify0to turn off angle correction.Note: Turning off angle correction is only safe to do if you know that your axial slices were acquired roughly orthogonal to the cord.
- Output files/folders:
csa_wm.csvandcsa_gm.csv: Two CSV files containing shape metrics for both the white and gray matter segmentations.