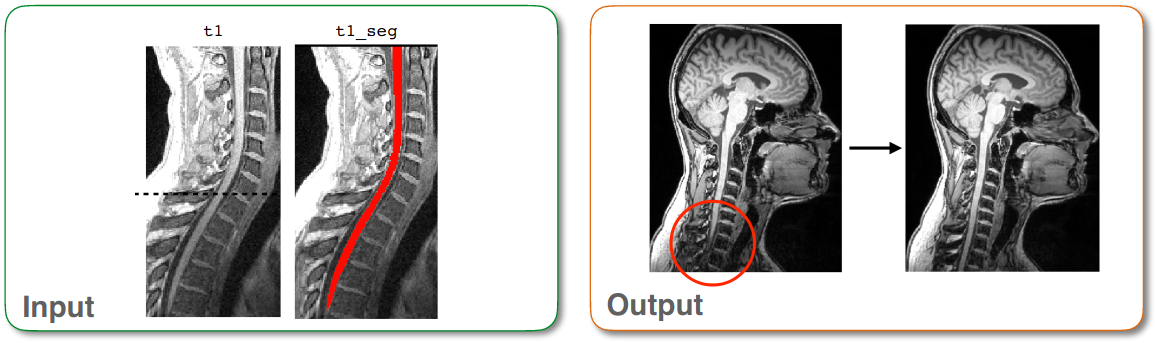
Visualizing misaligned cords with 2D sagittal flattening¶
Because some subjects (especially those with scoliosis) might not be perfectly aligned in the medial plane, it can sometimes be difficult to view the spinal cord when inspecting a subject’s images using individual 2D slices. To address this problem, the function sct_flatten_sagittal applies slice-wise deformation to align the cord in the medial plane.
Downloading the files for this tutorial¶
First, make sure that you have the following files in your working directory:
single_subject/data/t1.nii.gz: An anatomical spinal cord scan in the T1 contrast.single_subject/data/t1_seg.nii.gz: 3D segmentation of the spinal cord, corresponding to the T1 image.You can get these files by downloading data_visualizing-misaligned-cords.zip.
Running the command¶
Next, open up your terminal and run the following command:
sct_flatten_sagittal -i t1.nii.gz -s t1_seg.nii.gz
- Input arguments:
-i: The input image.-s: A spinal cord segmentation mask corresponding to the input image. This is needed assct_flatten_sagittaluses the centerline of the cord to align the image in the medial plane.
- Output files/folders:
t1_flatten.nii.gz: The input image, transformed in a way that ensures that the center medial (L-R) slice depicts the entire spinal cord.
Note
This process should be used for visualization purposes only, as it does not preserve the internal structure of the cord. If you would like to properly align the cord along the RL and AP direction for analysis, we recommend you use sct_straighten_spinalcord instead, as that transformation is reversible.