Preprocessing steps to highlight the spinal cord#
Prior to motion correction, it’s often helpful to crop dMRI data around the region of interest (e.g. the spinal cord). Doing so will speed up subsequent processing steps while also improving their accuracy.
To crop the data, we use the multi-step process below:
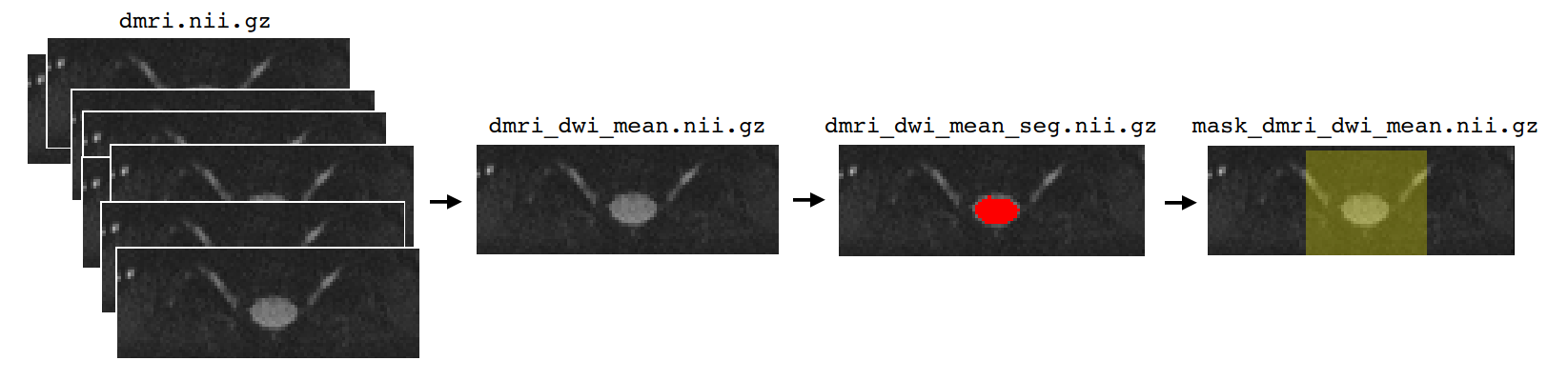
Generating a mean image#
First we generate the mean of the 4D dMRI data across the time axis in order to obtain a coarse 3D approximation of the image. This step is necessary because SCT’s spinal cord segmentation tools are designed for individual 3D volumes, rather than 4D images.
sct_dmri_separate_b0_and_dwi -i dmri.nii.gz -bvec bvecs.txt
- Input arguments:
-i: The input image.-bvec: The b-vector file used to separate DWI volumes from b0 volumes.
- Output files/folders:
dmri_dwi_mean.nii.gz: A 3D image containing the mean of the individual DWI volumes in the 4D dMRI image. This is the file we care most about, however the script will also output a b0 mean file, as well as 4D files containing the separated DWI and b0 volumes.
Generating a spinal cord segmentation#
The resulting 3D volume can then be used to obtain a cord segmentation.
sct_deepseg_sc -i dmri_dwi_mean.nii.gz -c dwi -qc ~/qc_singleSubj
- Input arguments:
-i: The input image.-c: The contrast of the input image.-qc: Directory for Quality Control reporting. QC reports allow us to evaluate the results slice-by-slice.
- Output files/folders:
dmri_dwi_mean_seg.nii.gz: An mask image containing the segmented spinal cord.
Note
Keep in mind that this segmentation was created from a non-motion-corrected mean dMRI image. While this segmentation is useful for coarsely highlighting the spinal cord, it won’t be applicable once motion correction is applied to the dMRI image, so you will need to re-run sct_deepseg_sc on the motion-corrected mean image later on.
Creating a mask around the spinal cord#
Once the segmentation is obtained, we can use it to create a mask around the cord.
sct_create_mask -i dmri_dwi_mean.nii.gz -p centerline,dmri_dwi_mean_seg.nii.gz -f cylinder -size 35mm
- Input arguments:
-i: The input image to create the mask from.-p: The process used to position the mask. Thecenterlineprocess will compute the center of mass for each slice ofdmri_dwi_mean_seg.nii.gz, then use those locations for the center of the mask at each slice.-f: The shape of the mask. Here, we create cylinder around the centerline.-size: The diameter of the mask.
- Output files/folders:
mask_dmri_dwi_mean.nii.gz: An imagine containing a mask surrounding the spinal cord.
This mask will be passed to the motion correction script so that the algorithm will process only the spinal cord region.