Applying the registration algorithm¶
To apply the registration algorithm, the following command is used:
sct_register_to_template -i t2_lumbar.nii.gz \
-s t2_lumbar_seg.nii.gz \
-ldisc t2_lumbar_labels.nii.gz \
-c t2 -qc ~/qc_singleSubj \
-param step=1,type=seg,algo=centermassrot:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,iter=3,slicewise=0:step=3,type=im,algo=syn,metric=CC,iter=3,slicewise=0
Note
If you get the following error message, it means that your installed version of the PAM50 template is too old:
ERROR: Wrong landmarks input. Labels must have correspondence in template space.
Label max provided: 60
Label max from template: 21
In that case, you should update it by following these instructions.
- Input arguments:
-i: Input image.-s: Segmented spinal cord corresponding to the input image.-ldisc: One or two labels located at posterior tip of the spinal cord discs. (In this case, we are using 1 disc label and 1 cauda equina label.)-c: Contrast of the image. Specifying this determines the version of the template to use. (Here,-c t2means thatPAM50_t2.nii.gzwill be used.)-qc: Directory for Quality Control reporting. QC reports allow us to evaluate the results slice-by-slice.-param: Registration parameters used to define the step-by-step refinements applied to the image. In this case, the parameters differ from the default registration parameters in the following ways: - Step 1 is specified astype=seg, to better rely on the lumbar segmentation. - A third ‘Syn’ step is added to fine-tune the registration and better match the cord shape.
- Output files/folders:
anat2template.nii.gz: The anatomical subject image (in this case,t2_lumbar.nii.gz) warped to the template space.template2anat.nii.gz: The template image (in this case,PAM50_t2.nii.gz) warped to the anatomical subject space.warp_anat2template.nii.gz: The 4D warping field that defines the transform from the anatomical image to the template image.warp_template2anat.nii.gz: The 4D warping field that defines the inverse transform from the template image to the anatomical image.
Once the command has finished, at the bottom of your terminal there will be instructions for inspecting the results using Quality Control (QC) reports. Optionally, If you have FSLeyes installed, a fsleyes command will printed as well.
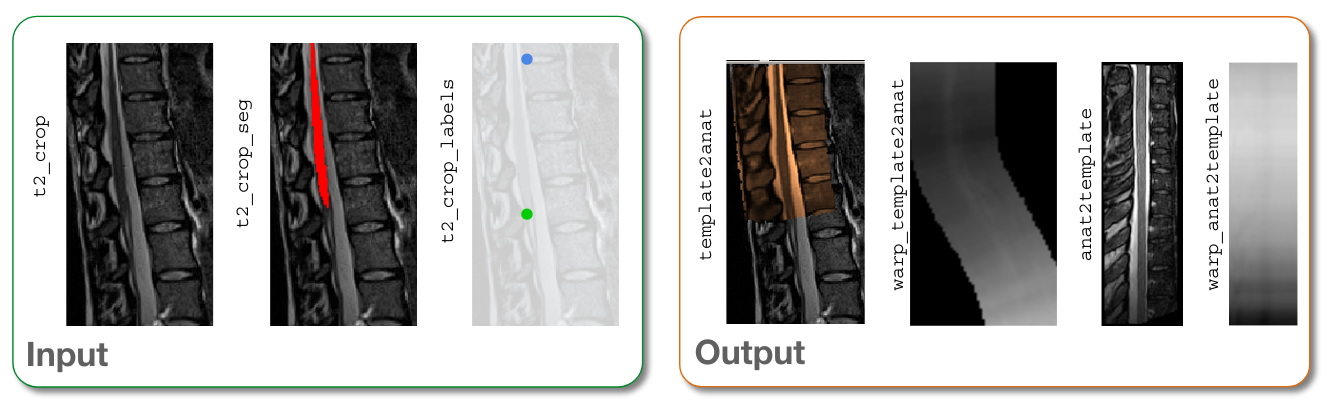
Input/output images after registration¶