Warping the spinal levels to the fMRI space¶
Now that we’ve registered the PAM50 template with the motion-corrected fMRI data, we can use the resulting warping field to transform the full template to the space of the dMRI data.
sct_warp_template -d fmri_moco_mean.nii.gz -w warp_template2fmri.nii.gz -a 0 -qc ~/qc_singleSubj
- Input arguments:
-d: Destination image the template will be warped to.-w: Warping field (template space to anatomical space).-a: Because-a 0is specified, the white and gray matter atlas will not be warped.-qc: Directory for Quality Control reporting. QC reports allow us to evaluate the results slice-by-slice.
- Output files/folders:
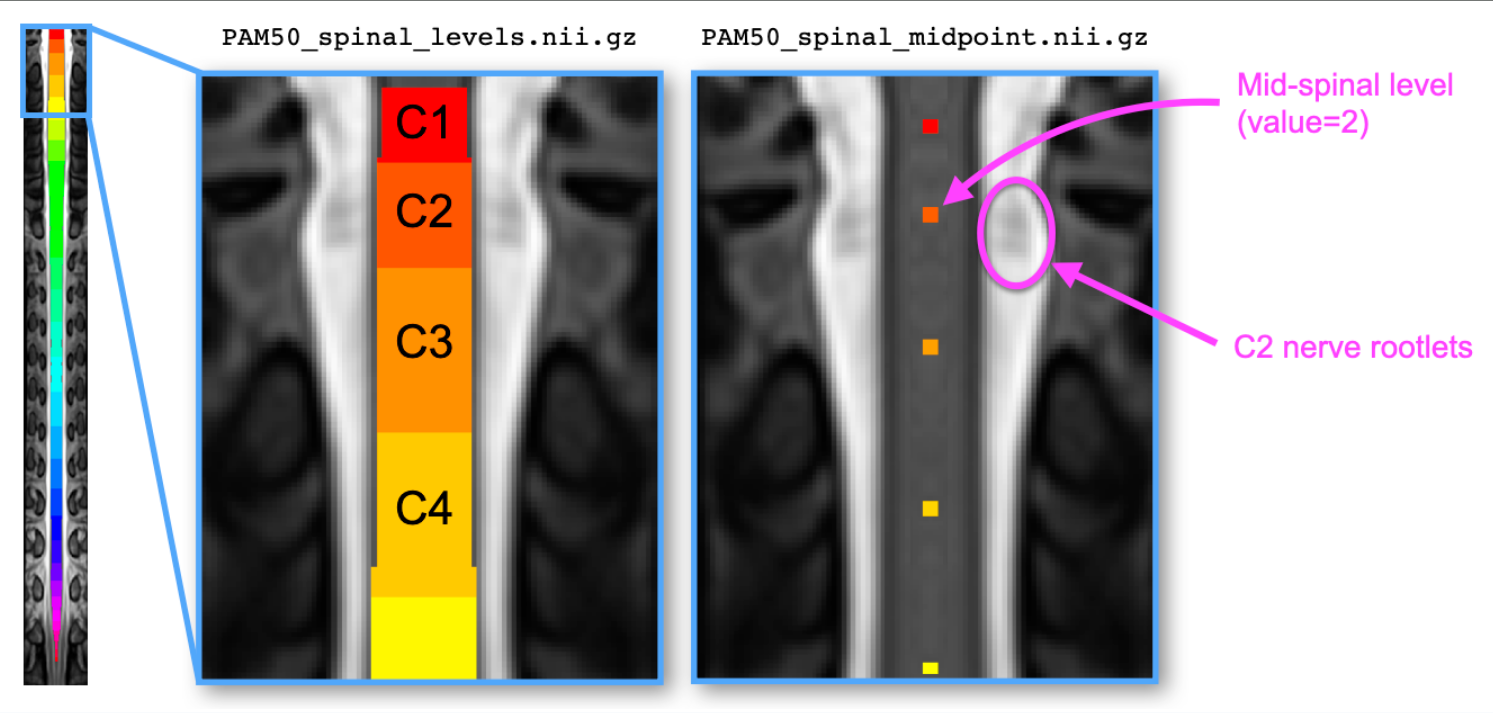
label/template/: This directory contains the entirety of the PAM50 template, transformed into the fMRI space. In particular, we care about thePAM50_spinal_levels.nii.gzandPAM50_spinal_midpoint.nii.gzfiles, which contain the spinal level “full body labels” and “single-voxel point labels” respectively.