Customizing the registration command¶
This page provides recommendations for how to adjust the registration commands if the default parameters are insufficient for your specific data and pipeline.
Note
Because choosing the right configuration for your data can be overwhelming, feel free to visit the SCT forum where you can ask for clarification and guidance.
The -param argument¶
The -param argument is used to specify the transformations that are applied at each step of the registration process. The easiest way to use -param is to start with the default values, and adjust one parameter at a time.
# Default values for '-param'. (Note: Normally, this would be written as one line, but a line break was added for readability.)
-param step=1,type=imseg,algo=centermassrot,metric=MeanSquares,iter=10,smooth=0,gradStep=0.5,slicewise=0,smoothWarpXY=2,pca_eigenratio_th=1.6:
step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,iter=3,smooth=1,gradStep=0.5,slicewise=0,smoothWarpXY=2,pca_eigenratio_th=1.6
Step 1 and step 2 can be modified, and additional steps can be added. In general, we recommend that you start with coarse-adjustment steps, then apply finer adjustments with each successive step.
-param “algo”¶
This parameter determines the type of nonrigid deformation to apply to the spinal cord at each step.
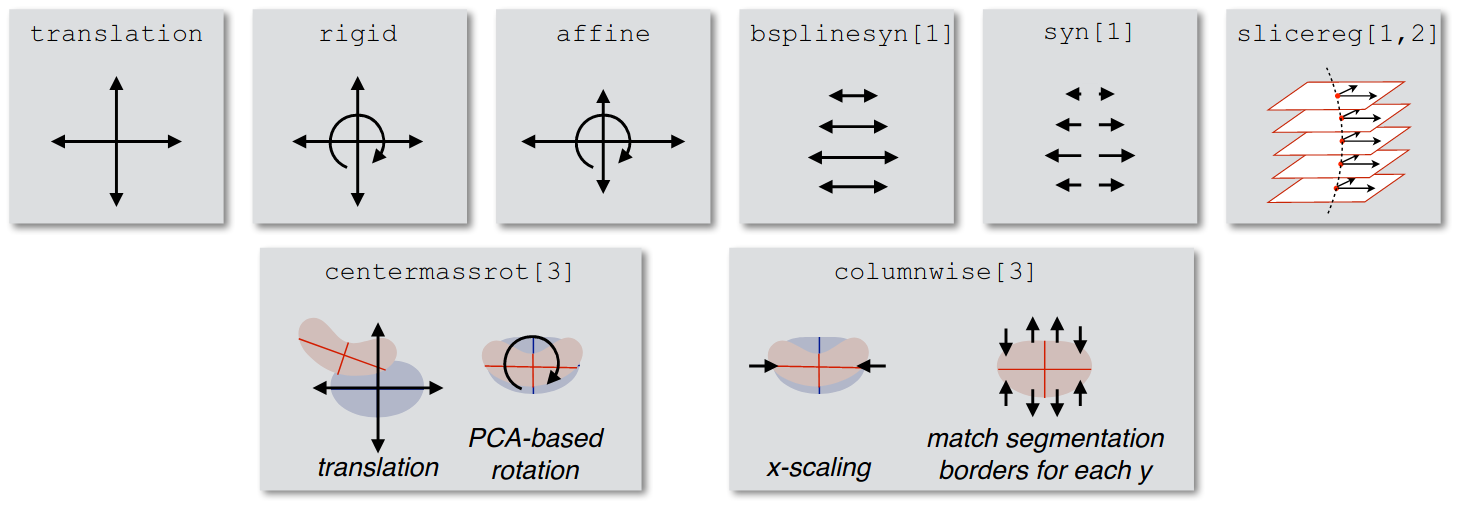
algo=translation:Axial (X-Y) translation.
algo=rigid:Axial (X-Y) translation + rotation about the Z axis.
algo=affine:Axial (X-Y) translation + rotation about the Z axis + axial (X-Y) scaling.
algo=syn:algo=b-splinesyn:B-Spline regularized form of SyN provided by ANTs.
algo=slicereg:Slice-by-slice axial (X-Y) translation, regularized along the Z axis. This can be used as both an initial alignment of a segmented cord centerline, or to align the centers of two images that are already close.
algo=centermassrot:An alignment of the center of mass of the segmented cord with the center of mass of the template. Similar to
algo=slicereg, but also includes rotation, to account for a turned cord due to e.g. compression on one side of the cord.algo=columnwise:This transformation involves a multi-step scaling operation with a much greater degree of freedom, so it is useful for highly compressed cords.
-param “type”¶
This parameter controls the type of data used for registration.
type=im:Use the input anatomical image.
type=seg:Use a separate segmentation image. Choose this if your image data contains distortions or artifacts, but you are confident in your segmentation. (For example, if you have manually corrected the segmentation.)
type=imseg:Use both the input image and a segmentation. Only for use with
algo=centermassrot.
-param “metric”¶
This parameter determines which similarity metric is used to match the vertebral levels between the anatomical image and the template.
metric=CC:Cross-correlation. Intensity-based comparison with a small amount of normalization. It is quite accurate, but takes longer to process compared to other approaches.
metric=MI:Mutual information. Entropy-based comparison. This method is faster overall, but requires large images to perform well. Cannot be used with
type=seg.metric=MeanSquares:Intensity-based comparison. Can only be used on images with the same intensity range. Should be used with
type=seg.
-param “slicewise”¶
This parameter controls whether or not transformations should be computed on a slice-by-slice basis:
slicewise=1:Apply deformations on a slice-by-slice basis.
slicewise=0:Regularize transformations across the Z axis.
The -ref argument¶
The flag -ref lets you select the destination for registration: either the template (default) or the subject’s native space.
-ref template:The subject will be registered to the template space, causing the anatomical image to be straightened. This method should be used by default.
-ref subject:With this setting, the template will be registered to the subject space (which does not require straightening). Use this approach if your image is acquired axially with highly anisotropic resolution (e.g. 0.7x0.7x5mm), because the straightening step can produce through-plane interpolation errors for thick slices.
The -ldisc argument¶
This argument is used to specify disc labels, rather than vertebral body labels. Vertebral body labels work well if you are only interested in a relatively small region (e.g. C2 —> C7). However, there are two main cases where you would want to instead use -ldisc:
- Large field of view:
If your volume spans a large superior-inferior length (e.g., C2 —> L1), the linear scaling between your subject and the template might produce inaccurate vertebral level matching between C2 and L1. In that case, you might prefer to rely on all intervertebral discs for a more accurate registration.
- Tiny field of view:
Conversely, if you have a very small field of view (e.g., covering only C3/C4), you can create a unique label at disc C3/C4 (value=4) and use
-ldiscfor registration. In that case, a single translation (no scaling) will be performed between the template and the subject.
Note
If more than 2 labels are provided, -ldisc is not compatible with -ref subject. For more information, please see the help: sct_register_to_template -h