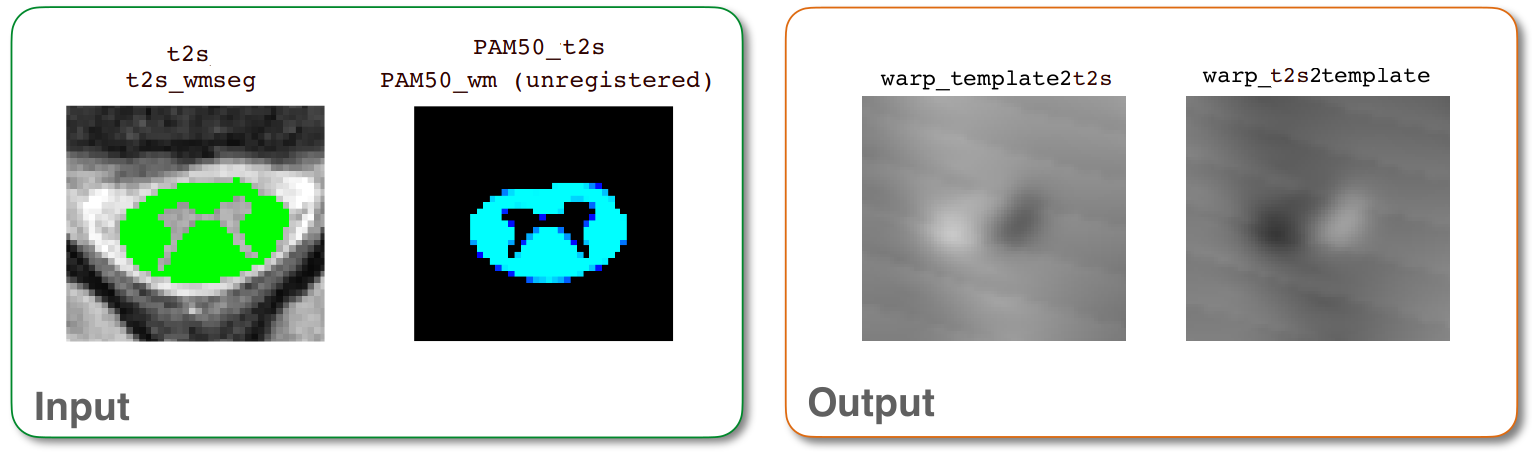
GM-informed registration between the PAM50 template and T2* data¶
Usually, template registration would be perfomed using the sct_register_to_template command. That command is important because it matches the vertebral levels of the data to that of the PAM50 template. Unfortunately, though, because T2* scans are typically acquired axially with thick slices, it is much more difficult to acquire the vertebral labels needed for the vertebral matching step.
To get around this limitation, we recommend that you first perform vertebral labeling and template registration using a different contrast for the same subject (e.g. T2 anatomical data, where vertebral levels are much more apparent). This will provide you with warping fields between the template and the data, which you can then re-use to initialize the T2* registration via the -initwarp and -initwarpinv flags. Doing so provides all of the benefits of vertebral matching, without having to label the T2* data directly.
Since we are starting the T2* registration with an initial transformation already applied, all that is left is fine-tuning for the T2* data. Here, we use a different command: sct_register_multimodal. This command is the more general, flexible counterpart to the sct_register_to_template command, as it provides more options to register any two images together.
sct_register_multimodal -i "${SCT_DIR}/data/PAM50/template/PAM50_t2s.nii.gz" \
-iseg "${SCT_DIR}/data/PAM50/template/PAM50_wm.nii.gz" \
-d t2s.nii.gz \
-dseg t2s_wmseg.nii.gz \
-initwarp ../t2/warp_template2anat.nii.gz \
-initwarpinv ../t2/warp_anat2template.nii.gz \
-owarp warp_template2t2s.nii.gz \
-owarpinv warp_t2s2template.nii.gz \
-param step=1,type=seg,algo=rigid:step=2,type=seg,metric=CC,algo=bsplinesyn,slicewise=1,iter=3:step=3,type=im,metric=CC,algo=syn,slicewise=1,iter=2 \
-qc ~/qc_singleSubj
- Input arguments:
-i: Source image. Here, we select the T2* version of the PAM50 template.-iseg: Segmentation for the source image. Here, we use the PAM50 segmented white matter volume, rather than the spinal cord volume. This allows us to account for both the cord and the gray matter shape.-d: Destination image.-dseg: Segmentation for the destination image. Here, we use the white matter segmentation for the same reasons as-iseg.-initwarp: Warping field used to initialize the source image. Here, we supply thewarp_template2anat.nii.gzfile from the previous T2 registration. (See: Registering labeled anatomical images to the PAM50 template)-initwarpinv: Warping field used to initialize the destination image. Here, we supply the inverse warping field,warp_anat2template.nii.gzfrom the previous T2 registration. (See: Registering labeled anatomical images to the PAM50 template)-param: Here, we will tweak the default registration parameters by specifying a different nonrigid deformation. The important change isalgo=rigid(rather thanalgo=centermass). The rigid algorithm is chosen because, by default, it is a volumetric transformation (i.e. the algorithm estimates a single rigid transformation for all slices). This makes it less sensitive to flaws in the segmentation compared to slicewise transformations, which is important when working with gray/white matter segmentations (as opposed to spinal cord segmentations).-owarp: The name of the output warping field. This is optional, and is only specified here to make the output filename a little clearer. By default, the filename would be automatically generated from the filenames-iand-d, which in this case would be the (less clear)warp_PAM50_t2s2t2s.nii.gz.-owarpinv: The name of the output inverse warping field. This is specified for the same reasons as-owarp.-qc: Directory for Quality Control reporting. QC reports allow us to evaluate the results slice-by-slice.
- Output files/folders:
PAM50_t2s_reg.nii.gz: The PAM50 template image, registered to the space of the T2s image.t2s_reg.nii.gx: The T2s image, registered to the space of the PAM50 template.warp_template2t2s.nii.gz: The warping field to transform the PAM50 template to the T2s space.warp_t2s2template.nii.gz: The warping field to transform the T2s image to the PAM50 template space.