Using sct_deepseg to segment the lumber region of the spinal cord¶
Note
Currently, sct_deepseg can only be used to segment the lumbar region of T2-weighted images. For other contrasts (T1w, T2*w, etc.), it is necessary to manually segment the lumbar region.
In this example, we begin with T2 anatomical scan of the lumbar region (t2_lumbar.nii.gz).
SCT’s lumbar segmentation tool works best if the lumbar region is the central feature of the image. So, it often helps to exclude the irrelevant portions of the spinal cord (brain, cervical region, and most of the thoracic region), as was done with this example scan.
Downloading the lumbar segmentation model¶
In previous registration tutorials, sct_deepseg_sc was used to segment the spinal cord. However, sct_deepseg_sc works best on the cervical and thoracic regions. To segment the lumbar region, we will need to use a different tool instead (sct_deepseg).
sct_deepseg is the more “general” cousin of sct_deepseg_sc, providing many different deep learning models for segmentation beyond just the spinal cord. To view the available models, run:
sct_deepseg -h
Then, to download and install the correct model, run:
sct_deepseg -install-task seg_lumbar_sc_t2w
Now, sct_deepseg can be used to segment the lumbar region of the spinal cord.
Segmenting the cropped image¶
Here, we simply feed the cropped image to the deep learning model to segment the lumbar region.
sct_deepseg -i t2_lumbar.nii.gz -task seg_lumbar_sc_t2w
- Input arguments:
-i: Input image-task: The deep learning segmentation task to apply to the image. In this case, we want seg_lumbar_sc_t2w.
- Output files/folders:
t2_lumbar_seg.nii.gz: 3D binary mask of the segmented spinal cord
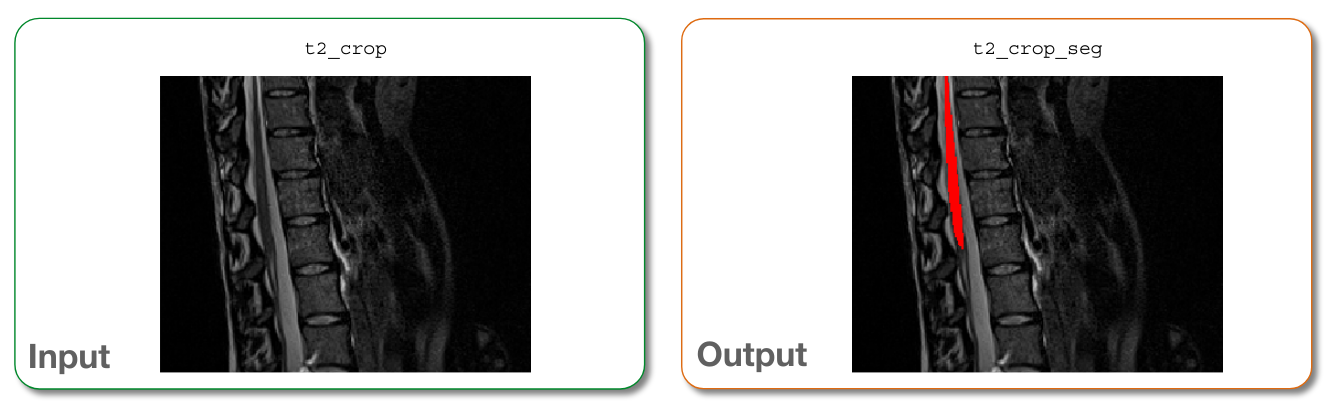
Input/output images after segmentation¶