Hands-on: Using sct_deepseg_sc on T2 data¶
Run the following command to process the image:
sct_deepseg_sc -i t2.nii.gz -c t2 -qc ~/qc_singleSubj
- Input arguments:
-i: Input image-c: Contrast of the input image-qc: Directory for Quality Control reporting. QC reports allow us to evaluate the segmentation slice-by-slice
- Output files/folders:
t2_seg.nii.gz: 3D binary mask of the segmented spinal cord
Once the command has finished, at the bottom of your terminal there will be instructions for inspecting the results using Quality Control (QC) reports. You may also simply refresh the webpage that was generated in the previous sections to see the new results.
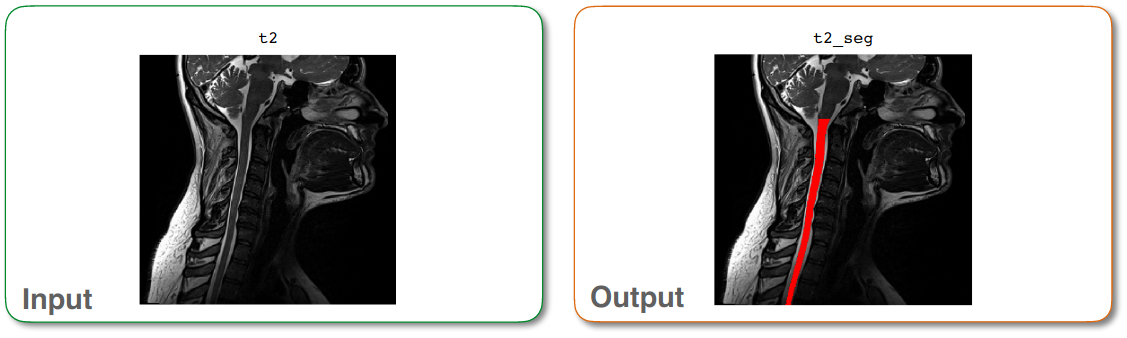
Output of sct_deepseg_sc¶